Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Chiba, Kaori*; Matsui, Takuro*; Chatake, Toshiyuki*; Ohara, Takashi; Tanaka, Ichiro*; Yutani, Katsuhide*; Niimura, Nobuo*
Protein Science, 32(10), p.e4765_1 - e4765_13, 2023/10
Times Cited Count:0 Percentile:0(Biochemistry & Molecular Biology) -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Noda, Hisanobu*; Tamada, Taro; Kazuma, Kohei*; Suzuki, Masahiko*; Blaber, M.; Kuroki, Ryota
Protein Science, 24(3), p.395 - 407, 2015/03
Times Cited Count:62 Percentile:88.58(Biochemistry & Molecular Biology)UDP-glucose: anthocyanidin 3- -glucosyltransferase (UGT78K6) from
-glucosyltransferase (UGT78K6) from  catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85
catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85  , 2.55
, 2.55  , 2.70
, 2.70  and 1.75
and 1.75  respectively. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180
respectively. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180 rotation about the O1-O3 axis of the flavonoid backbones observed in
rotation about the O1-O3 axis of the flavonoid backbones observed in 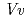 GT1 and UGT78G1. These substrate recognition schemes suggest the potential for controlled synthesis of natural pigments.
GT1 and UGT78G1. These substrate recognition schemes suggest the potential for controlled synthesis of natural pigments.

Adachi, Motoyasu; Hirayama, Hiroshi; Shimizu, Rumi; Sato, Katsuya; Narumi, Issey*; Kuroki, Ryota
Protein Science, 23(10), p.1349 - 1358, 2014/10
Times Cited Count:9 Percentile:25.51(Biochemistry & Molecular Biology)Pleiotropic protein promoting DNA repair A (PprA) is a key protein that facilitates the extreme radioresistance of  . To clarify the role of PprA in the radioresistance mechanism, the interaction between recombinant PprA expressed in Escherichia coli with several double-stranded DNAs was investigated. In a gel-shift assay, the band shift of supercoiled pUC19 DNA caused by the binding of PprA showed a bimodal distribution, which was promoted by the addition of 1 mM Mg, Ca, or Sr ions. The dissociation constant of the PprA-supercoiled pUC19 DNA complex, calculated from the relative portions of shifted bands, was 0.6
. To clarify the role of PprA in the radioresistance mechanism, the interaction between recombinant PprA expressed in Escherichia coli with several double-stranded DNAs was investigated. In a gel-shift assay, the band shift of supercoiled pUC19 DNA caused by the binding of PprA showed a bimodal distribution, which was promoted by the addition of 1 mM Mg, Ca, or Sr ions. The dissociation constant of the PprA-supercoiled pUC19 DNA complex, calculated from the relative portions of shifted bands, was 0.6 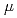 M with a Hill coefficient of 3.3 in the presence of 1 mM Mg acetate. This indicates that at least 281 PprA molecules are required to saturate a supercoiled pUC19 DNA, which is consistent with the number of bound PprA molecules estimated by the UV absorption of the PprA-pUC19 complex purified by gel filtration. This saturation also suggests linear polymerization of PprA along the dsDNA. On the other hand, the bands of linear dsDNA and nicked circular dsDNA that eventually formed PprA complexes did not saturate, but created larger molecular complexes when the PprA concentration was greater than 1.3
M with a Hill coefficient of 3.3 in the presence of 1 mM Mg acetate. This indicates that at least 281 PprA molecules are required to saturate a supercoiled pUC19 DNA, which is consistent with the number of bound PprA molecules estimated by the UV absorption of the PprA-pUC19 complex purified by gel filtration. This saturation also suggests linear polymerization of PprA along the dsDNA. On the other hand, the bands of linear dsDNA and nicked circular dsDNA that eventually formed PprA complexes did not saturate, but created larger molecular complexes when the PprA concentration was greater than 1.3 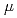 M. This result implies that DNA-bound PprA aids association of the termini of damaged DNAs, which is regulated by the concentration of PprA.
M. This result implies that DNA-bound PprA aids association of the termini of damaged DNAs, which is regulated by the concentration of PprA.
 KM1
KM1Okazaki, Nobuo; Tamada, Taro; Feese, M. D.*; Kato, Masaru*; Miura, Yutaka*; Komeda, Toshihiro*; Kobayashi, Kazuo*; Kondo, Keiji*; Blaber, M.*; Kuroki, Ryota
Protein Science, 21(4), p.539 - 552, 2012/04
Times Cited Count:3 Percentile:6.67(Biochemistry & Molecular Biology) sp. nucleoside diphosphate kinase
sp. nucleoside diphosphate kinaseArai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Matsumoto, Fumiko; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Protein Science, 21(4), p.498 - 510, 2012/04
Times Cited Count:14 Percentile:34.5(Biochemistry & Molecular Biology)In order to clarify the oligomer state of nucleoside diphosphate kinase (NDK) from moderately halophilic  sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
Yamada, Hidenori*; Tamada, Taro; Kosaka, Megumi*; Miyata, Kohei*; Fujiki, Shinya*; Tano, Masaru*; Moriya, Masayuki*; Yamanishi, Mamoru*; Honjo, Eijiro; Tada, Horiko*; et al.
Protein Science, 16(7), p.1389 - 1397, 2007/07
Times Cited Count:39 Percentile:59.61(Biochemistry & Molecular Biology)In an attempt to control protein incorporation in a crystal lattice, a leucine zipper-like hydrophobic interface (comprising four leucine residues) was introduced into a helical region (helix 2) of the human pancreatic ribonuclease 1 (RNase 1) that was predicted to form a suitable crystallization interface. Although crystallization of wild type RNase 1 has not yet been reported, the RNase 1 mutant having four leucines (4L-RNase 1) was successfully crystallized under several different conditions. The crystal structures were subsequently determined by X-ray crystallography by molecular replacement using the structure of bovine RNase A. The overall structure of 4L-RNase 1 is quite similar to that of the bovine RNase A, and the introduced leucine residues formed the designed crystal interface. To further characterize the role of the introduced leucine residues in crystallization of RNase 1, the number of leucines was reduced to three or two (3L- and 2L-RNase 1, respectively). Both mutants crystallized and a similar hydrophobic interface as in 4L-RNase 1 was observed. A related approach to engineer crystal contacts at helix 3 of RNase 1 (N4L-RNase 1) was also evaluated. N4L-RNase 1 also successfully crystallized, and formed the expected hydrophobic packing interface. These results suggest that appropriate introduction of a leucine zipper-like hydrophobic interface can promote intra molecular symmetry for more efficient protein crystallization in crystal lattice engineering efforts.
Matsuda, Keiko*; Nishioka, Takaaki*; Kinoshita, Kengo*; Kawabata, Takeshi*; Go, Nobuhiro
Protein Science, 12(10), p.2239 - 2251, 2003/10
Times Cited Count:10 Percentile:17.1(Biochemistry & Molecular Biology)no abstracts in English